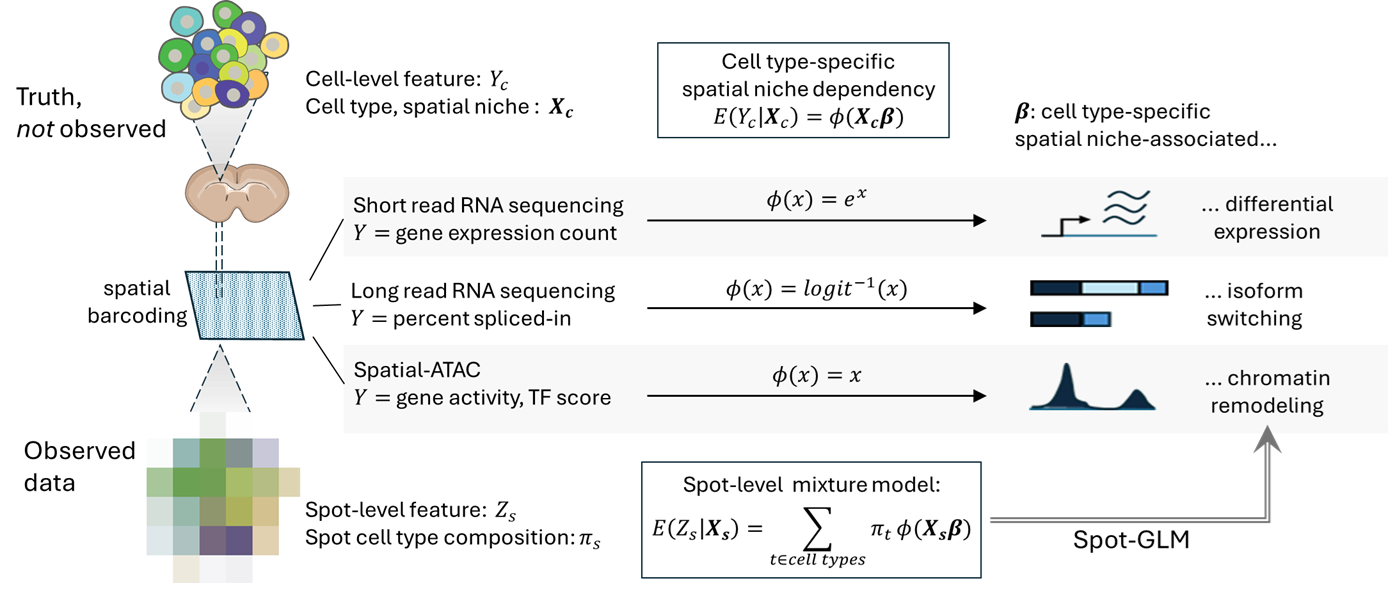
SpotGLM is an R package for performing cell type-specific differential testing in spatial omics data. It adapts to different data modalities, such as gene expression, chromatin accessibility, or isoform usage, and identifies cell type specific changes associated with local tissue context. SpotGLM accounts for the mixed-cell composition inherent to spatial barcoding technologies.
SpotGLM is integrated with the SPARROW package, making it fast and ultra-scalable for data sets with millions of spots.
Installation
To install SpotGLM from GitHub:
install.packages("devtools")
devtools::install_github("kaishumason/SpotGLM") # installWe strongly recommend using SpotGLM with SPARROW, which is a power-preserving data reduction method which can speed up analyses. SPARROW can also be installed from GitHUB:
install.packages("devtools")
devtools::install_github("kaishumason/SPARROW") # installGetting Started
Please follow these tutorials to get started on your data:
Using SpotGLM with SPARROW on Visium HD
SpotGLM for isoform switching analysis on spatial long read data
SpotGLM for spatial ATAC analysis
Here is a more technical tutorial that applies spotGLM to simulation data. By working with simulation data, you can see the generative model used in spotGLM: